Paquetes
library(tidyverse)
library(lubridate)
library(gganimate)
library(png)
library(gifski)
library(readxl)
Dataset
df <- read_csv(url('https://covid.ourworldindata.org/data/ecdc/full_data.csv'))
## Parsed with column specification:
## cols(
## date = col_date(format = ""),
## location = col_character(),
## new_cases = col_double(),
## new_deaths = col_double(),
## total_cases = col_double(),
## total_deaths = col_double(),
## weekly_cases = col_double(),
## weekly_deaths = col_double(),
## biweekly_cases = col_double(),
## biweekly_deaths = col_double()
## )
Total de casos confirmados y fallecimientos
Número total (acumulado) de casos confirmados y fallecimientos en el mundo.
df_plot <- df %>%
filter(location == "World") %>%
select(date, location, total_cases, total_deaths) %>%
gather(key = "variable", value = "cantidad", 3:4)
df_plot_animated <- df_plot %>%
ggplot(aes(x = variable,
y = cantidad,
fill = variable)) +
geom_bar(stat= "identity",
color = "white",
show.legend = FALSE) +
geom_text(aes(label = as.numeric(cantidad)),
position = position_dodge(0.9),
vjust= -1.2,
size = 5,
color = "darkgrey")+
scale_y_continuous(labels = scales::comma) +
scale_fill_manual(values = c("orange", "darkgrey")) +
theme_minimal()+
enter_appear() +
transition_states(date,
transition_length = 1,
state_length = 1) +
labs(title = "COVID-19: Contagiados y fallecimientos (acumulado) \n Fecha: {closest_state}",
subtitle = "",
y = "",
x = "",
caption = "Fuente: https://covid.ourworldindata.org") +
theme(panel.grid.major.x = element_blank(),
plot.title = element_text(size = 18,
face = "bold",
hjust = 0.5),
plot.subtitle = element_text(size=14,
face = "plain",
hjust = 0.5))
animate(df_plot_animated, nframes= 380, width = 600, height= 600, fps = 7)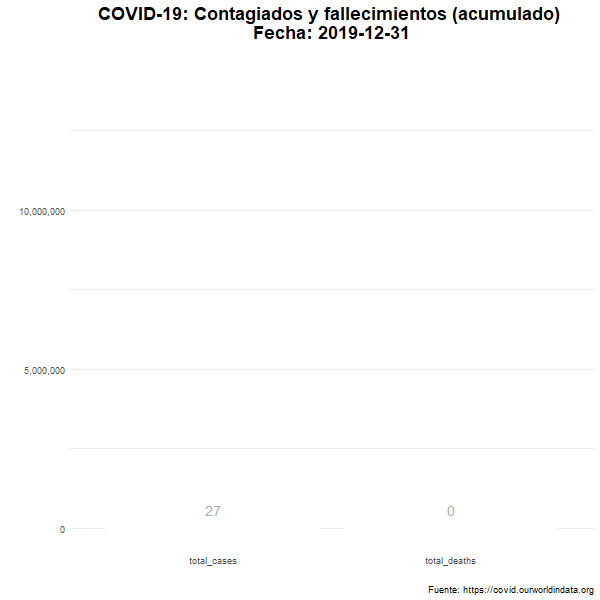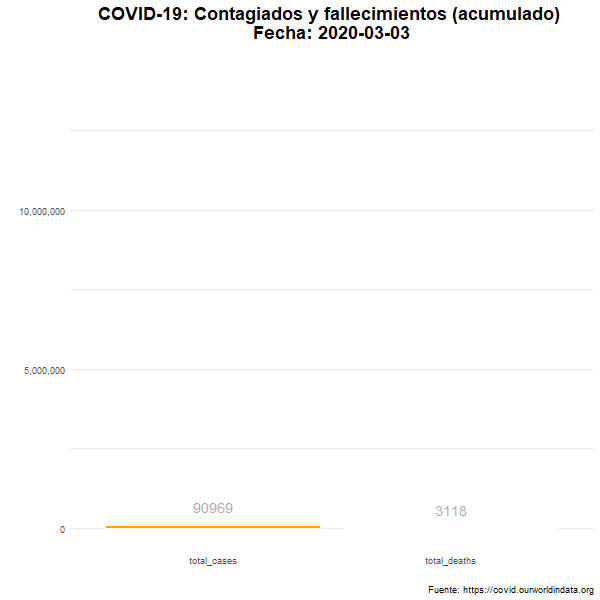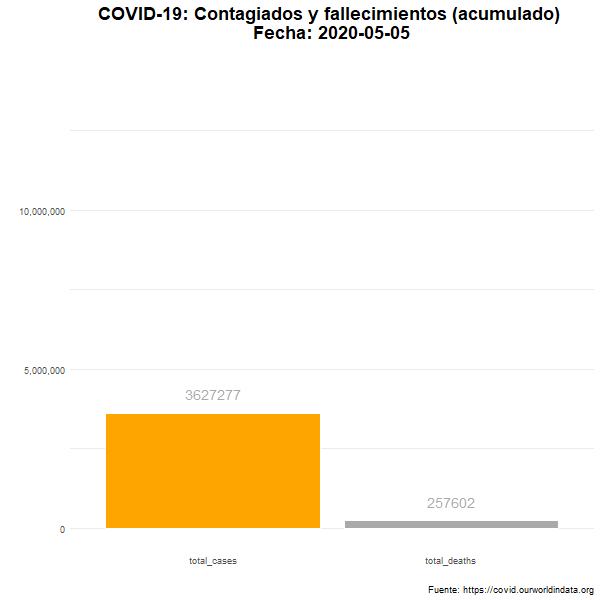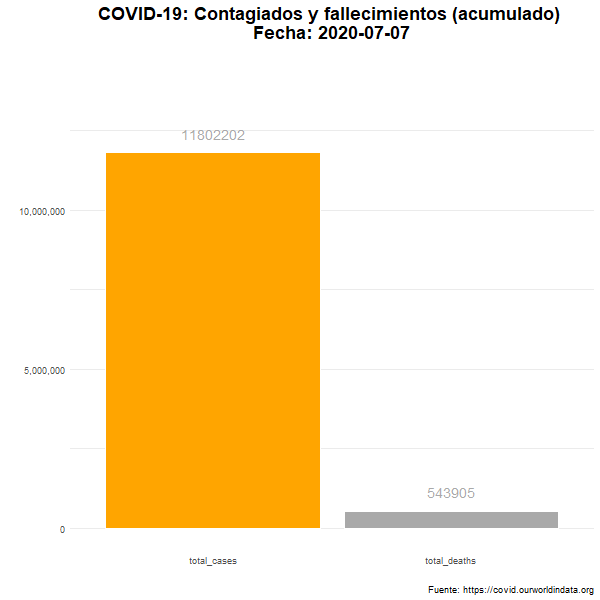
anim_save("df_plot_animated.gif")
Total de nuevos casos y nuevos fallecidos
Número diario de casos confirmados y fallecimientos en el mundo.
df_plot_new <- df %>%
filter(location == "World") %>%
select(date, location, new_cases, new_deaths) %>%
gather(key = "variable", value = "cantidad", 3:4)
df_plot_new_animated <- df_plot_new %>%
ggplot(aes(x = variable,
y = cantidad,
fill = variable)) +
geom_bar(stat= "identity",
color = "white",
show.legend = FALSE) +
geom_text(aes(label = as.numeric(cantidad)),
position = position_dodge(0.9),
vjust= -1.2,
size = 5,
color = "darkgrey")+
scale_y_continuous(labels = scales::comma) +
scale_fill_manual(values = c("orange", "darkgrey")) +
theme_minimal()+
enter_appear() +
transition_states(date,
transition_length = 1,
state_length = 1) +
labs(title = "COVID-19: Contagiados y fallecimientos (diario) \n Fecha {closest_state}",
subtitle = "",
y = "",
x = "",
caption = "Fuente: https://covid.ourworldindata.org") +
theme(panel.grid.major.x = element_blank(),
plot.title = element_text(size = 18,
face = "bold",
hjust = 0.5),
plot.subtitle = element_text(size=14,
face = "plain",
hjust = 0.5))
animate(df_plot_new_animated, nframes= 390, width = 600, height= 600, fps = 7)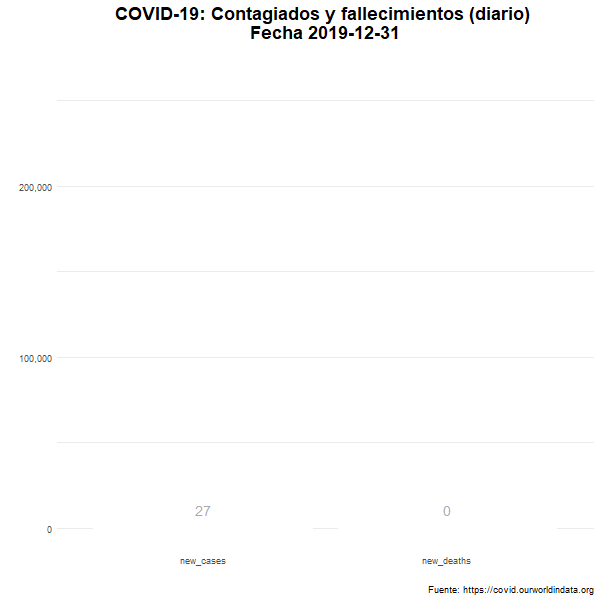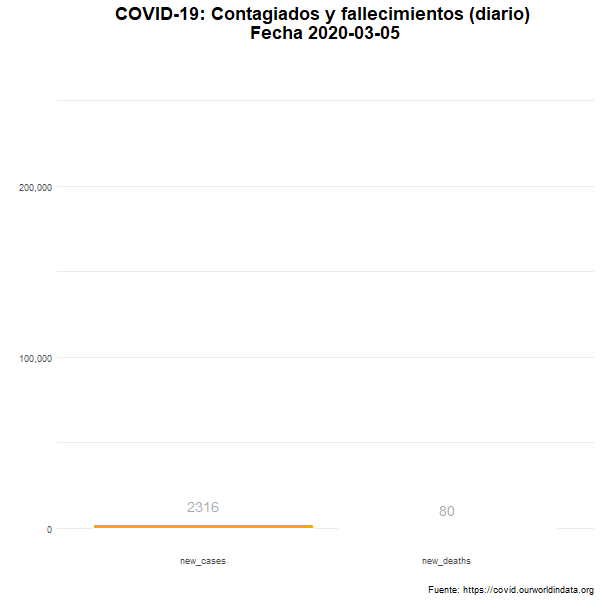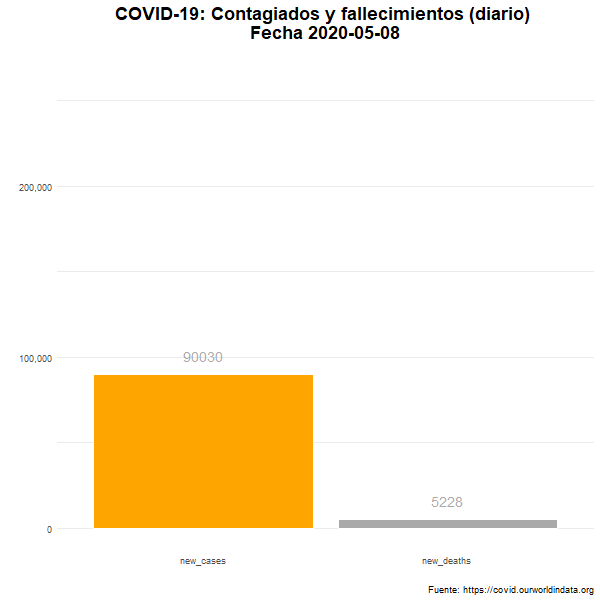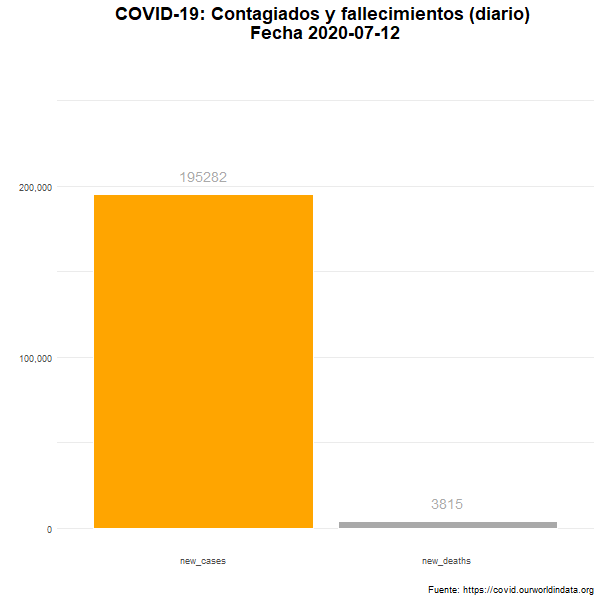
anim_save("df_plot_new_animated.gif")
Evolución del número de nuevos casos confirmados
Evolución temporal del número de personas contagiadas por COVID-19 en el mundo.
df_plot_new_trans <- df %>%
filter(location == "World") %>%
select(date, location, new_cases) %>%
group_by(date)
df_plot_new_transition <- df_plot_new_trans %>%
group_by(date) %>%
ggplot(aes(x = date,
y = new_cases,
fill = "orange")) +
geom_bar(stat= "identity",
color = "white",
show.legend = FALSE) +
scale_y_continuous(labels = scales::comma) +
theme_minimal()+
enter_appear() +
transition_states(date) +
shadow_mark() +
enter_grow() +
enter_fade() +
labs(title = "COVID-19: Contagiados (diario) \n Fecha: {closest_state}",
subtitle = "",
y = "",
x = "",
caption = "Fuente: https://covid.ourworldindata.org") +
theme(panel.grid.major.x = element_blank(),
plot.title = element_text(size = 18,
face = "bold",
hjust = 0.5),
plot.subtitle = element_text(size=14,
face = "plain",
hjust = 0.5))
animate(df_plot_new_transition, nframes= 390, width = 1000, height= 600, fps = 7)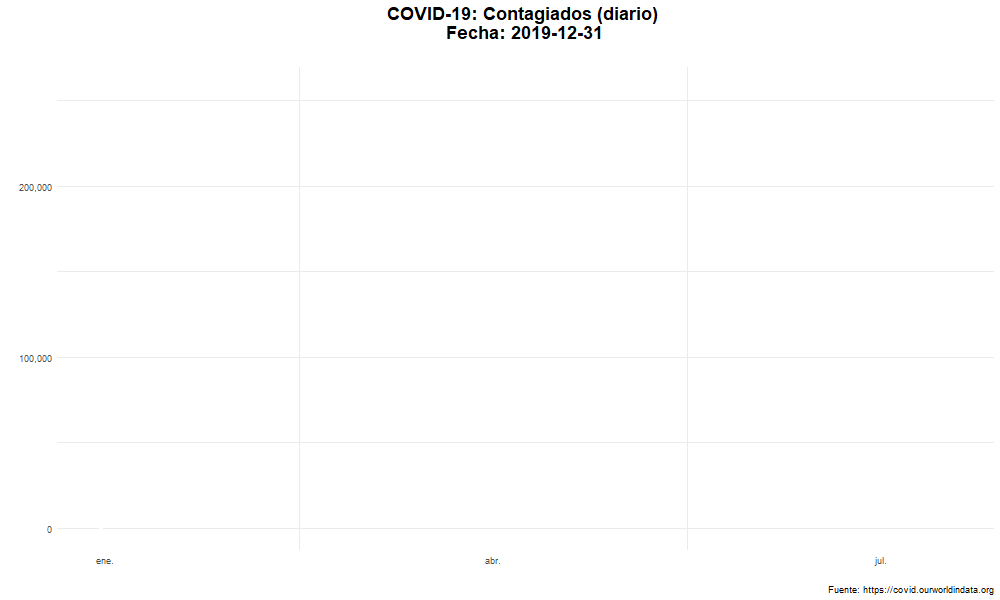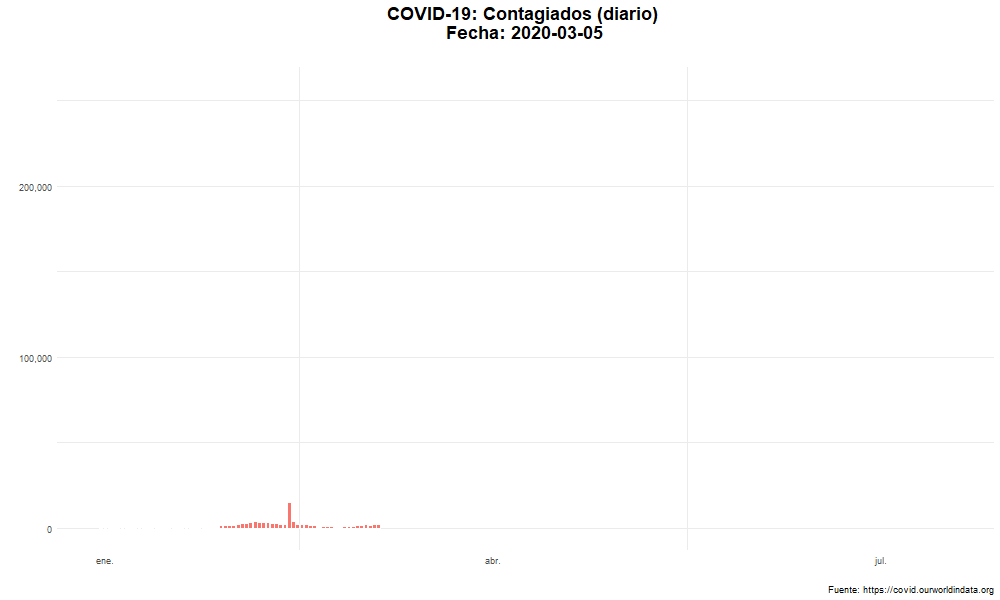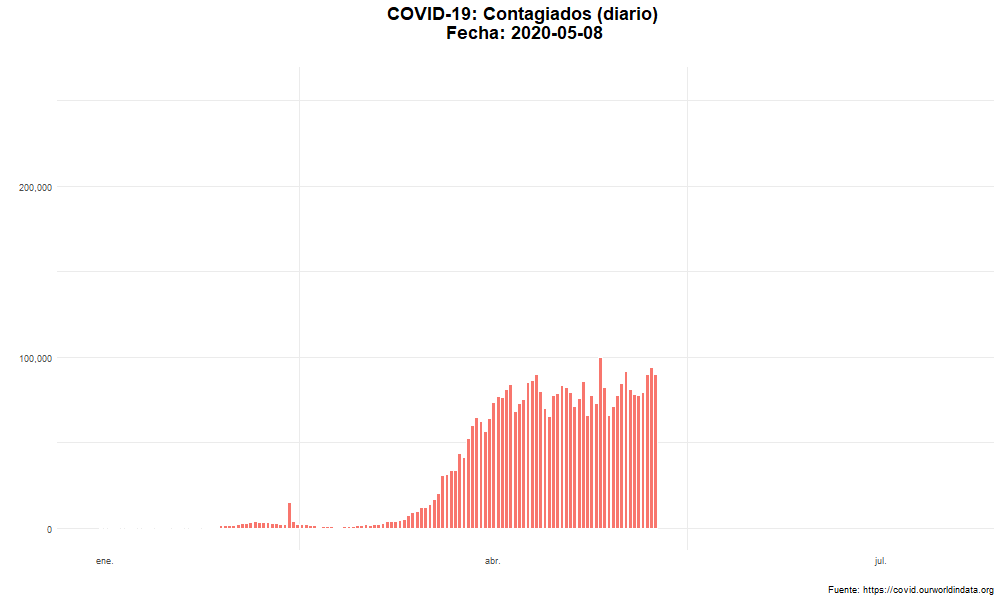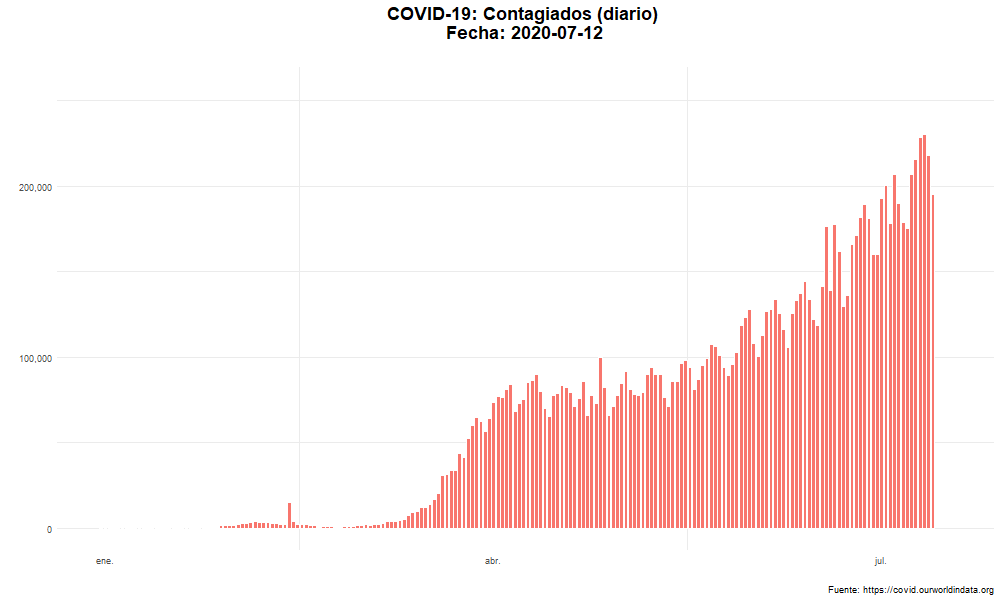
anim_save("df_plot_new_transition.gif")